| The basic ideas on crossdating are described in the section Dendrochronology, curve matching and mathematics. In this section we will look more on the necessary conditions and methods for finding the correct crossdating point between two curves. |
| A basic idea in crossdating is that ring width curves from different trees can be matched towards each other. So if the felling year for one of the trees is known, then the felling year for the other tree can be found by matching the curves as visualized in the diagram above. Figure 1 shows two curves which match perfectly towards each other. Now, this perfect matching is never the case in an actual situation with curves from two trees. This makes actual matching more difficult than understood by figure 1. |
|
A "correlation coefficient value" can be calculated out of a comparison of two curves which overlap each other at a certain position. The result is a value between -1 and 1, where 1 corresponds to a perfect match and other values above 0 represent more or less good matches. The correlation coefficient was described in the section Dendrochronology, curve matching and mathematics. After calculating a correlation coefficient value for every possible overlapping position between two curves we can then sort out the best matching points. To make us convinced that the best matching point of them represents the correct match, its correlation coefficient value has to be high enough and calculated for a sufficiently long overlap between the curves. We also require that the best matching point should be clearly discriminated, i.e. there should be no competing alternatives. We will now look at some crossdating cases to make us better understand under what conditions the correlation coefficient can be used for crossdating. |
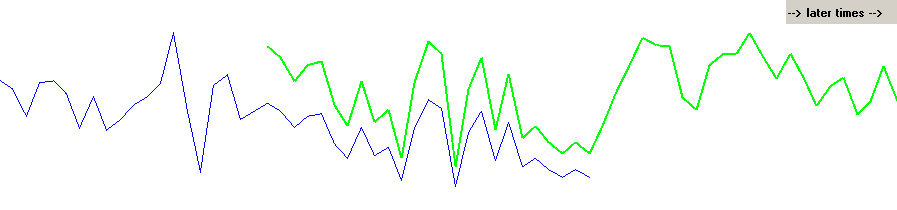
| It is common that tree rings grow wider when a tree is young which makes it difficult to compare its growth curve to a mean value curve. Though we are actually looking at the correct match in the figure 2 above, its corresponding correlation coefficient value (CC) is only 0.10 When looking at all possible matching positions, that correct matching point will not stand out. I.e. it will not be discriminated and found because of all the false matches with higher CC values!. |
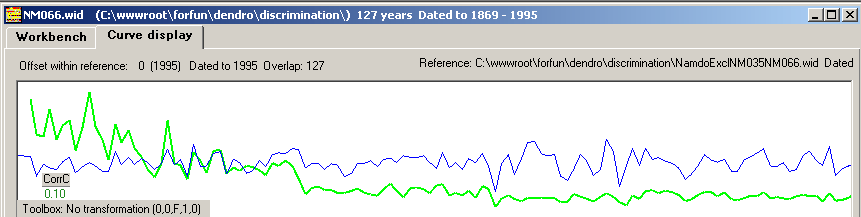
The correct match stands out with a high CC value at 1995.
|
This case can be handled by "detrending" the ring width growth curve (as shown in figure 3) before we do the correlation coefficient analysis. This will
make the correct matching point stand out properly (figure 4)! I.e. when comparing pure ring widths of two curves, detrending is a necessity!
Note: In contrast to this, detrending is NOT a first necessary step if we prepare the ring width curves by normalizing them before doing the correlation analysis. Normalization can be done with e.g. the Baillie/Pilcher or the P2Yrs methods, see below and it implies a type of detrending. |
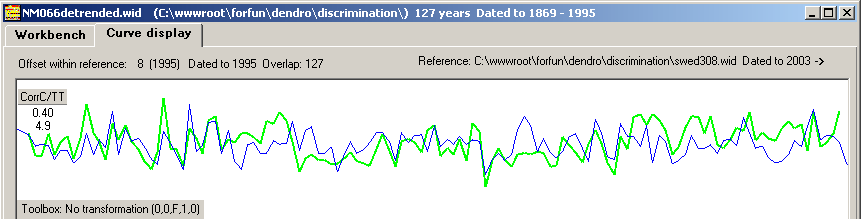
The correlation coefficient is only 0.40
| In a case when the ring width curve is compared to a reference from a more distant place, comparing curves becomes more difficult - like in figure 5 above where we compare our curve from the island of Nämdö with a mean value curve from Saltsjöbaden on the mainland some 25 km away. Then detrending is not a sufficient method to make the curves look alike! |
Transforming curves with normalization methods before we compare them.
| To make curve matching and crossdating based on correlation coefficient (CC) values more efficient we have to use more powerful transformation methods than just detrending for preparing ring width data. By dividing each ring width value with the mean of neighbour ring widths, we remove the strong dependence between adjacent rings. In CDendro these transformations are named normalization. |
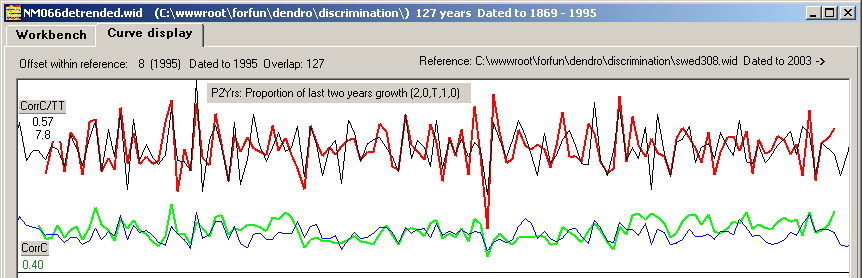
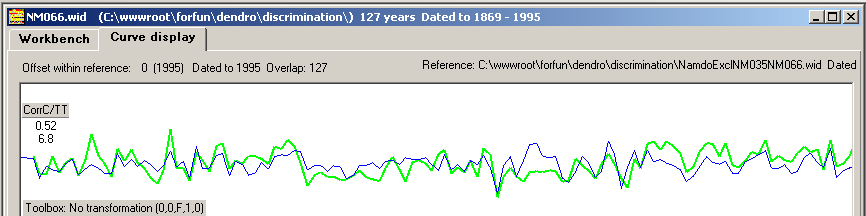
The upper curves (red/black) are transformed from ring width curves using the "Proportion of last two years growth, P2Yrs" normalization method.
The normalized curves match each other with high correlation values!
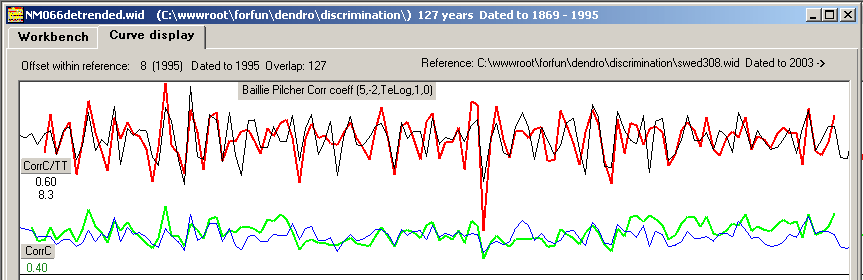
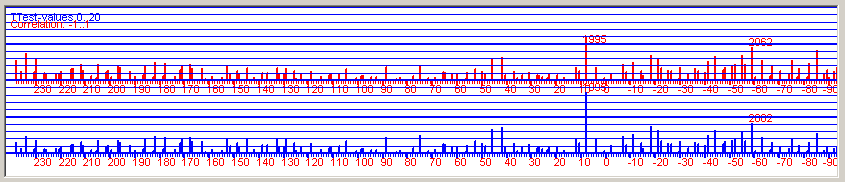
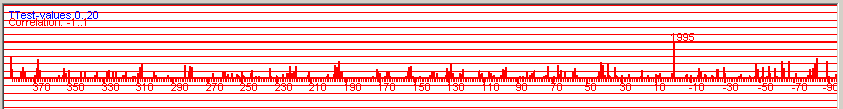
| Figure 8. Positive correlation coefficients and T-values for all possible overlapping positions when crossdating the detrended and normalized NM066 curve towards a normalized mean value curve from Saltsjöbaden on the mainland, some 25 km distance from the island of Nämdö. Baillie/Pilcher normalization. Note: The diagram was created with the detrended version of NM066, but the diagram will look almost exactly the same when using the original undetrended NM066, i.e. detrending is NOT a first necessary step before crossdating when using CDendro normalization as the normalization algorithms include sort of a detrending method. (But detrending is necessary before building mean value curves!) |
|
There are a number of different normalization methods that can be used.
Figure 6 and 7 show normalized curves based on the P2Yrs method and on the Baillie/Pilcher method. The green/blue curves show
the detrended NM066 ring width curve matched towards the mean value ring width curve with CC=0.40 The red/black curves are based on normalized values
giving much higher CC values. With both normalization methods the match becomes not only clear but also properly discriminated as shown in the pile diagram above
based on the Baillie/Pilcher method: There is no competing match! (Figure 8).
The "Proportion of last two years growth, P2Yrs" method divides each ring width with the width of the two last years, i.e. the normalized value d(y) = w(y)/(w(y)+w(y-1)) where w(y) is the ring width of year y and w(y-1) is that of the previous year. The algorithm for the Baillie/Pilcher method is: d(y) = eLog( 5* w(y)/( w(y-2)+ w(y-1) + w(y) + w(y+1) + w(y+2) )) You will find details about these algorithms under the menu command "Settings/Toolbox for normalization algorithms" in CDendro. There is also a section on this on our wiki: Cybis dendro wiki on normalization In the section What is a good TTest value to ensure a dating? you will find a comparison of these methods and their error rate for various "Lowest required T-value" (selected safety level for a match). |
|
After calculating a correlation coefficient for every possible overlapping position between two curves we can sort out the positions where the curves best match each other.
To make us convinced that the best matching position of them represents the correct match, its correlation coefficient value has to be high enough and calculated for a sufficiently
long overlap between the curves.
We also require that the best matching position should be clearly discriminated, i.e. there should be no competing alternatives, but instead a good margin to the "next best match".
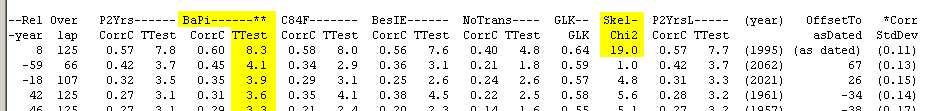
Figure 8 above, shows an example of good discrimination where the best matching position has a T-value above 8 and the next best match is at T=4.1 This matching can also be described with a table on the CDendro "Workbench" where correlation coefficients or T-values can be sorted in decreasing order, see figure 9. Please note the big difference in T-value between the topmost row and the row below it! |
|
When there is a very narrow ring within a sample and another very narrow ring within a reference curve (or another sample used for crossdating) then several normalization algorithms have a strong tendency to allow these rings to attract each other. This is because the overall shape of the curves are dominated by the look of these down pointing peaks. Our practical experience shows that the "Skeleton Chi2" algorithm has yet never been fooled by such a case, so it should always be checked before you agree to a matching proposal in CDendro. There is a separate section on this matter at How to get fooled by your normalization method and some too narrow ring widths. To avoid this problem with "narrow rings attracting each other", it might be best in CDendro to alway use the P2YrsL method for sorting out the best match. With this method standard deviations for normalized data are calculated and the final normalized curve has its peaks cut at a level corresponding to 2.6 * the standard deviation. (The 2.6 level is settable.) This makes the downward peaks less extreme which also lessens the risk of fooling CDendro. |
The auto-correlation function - cross correlating a signal towards itself
|
We will now again use the correlation coefficients to sort out the best and hopefully correct match between two ring width curves. To make this
case somewhat extreme, we will compare the curve towards an exact copy of itself!
For your visual comparison of two copies of the same ring with curve, this is a very simple and obvious case: Just put the end years over each other!
Though if there were an unknown number of years missing at both ends of a long curve, how would you then find the match?
How does the correlation coefficient behave when testing all possible overlapping positions for this case of matching two identical curves? |
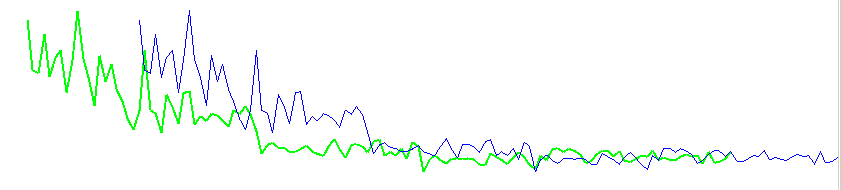
| For each possible overlapping position with at least e.g. 30 overlapping years, we will calculate a correlation coefficient value. We then plot these numbers, side by side as narrow columns where a column height corresponds to the correlation coefficient value, see figure 11. |
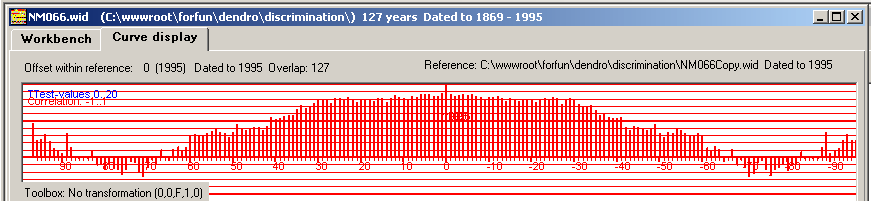
|
When the two identical curves lay exactly over each other as they should for an exact match, we get a correlation coefficient value of "+1".
This corresponds to the top in the middle of the diagram. The curve above is called "the autocorrelation function" for this ring width curve.
There are many points around the correct matching point that give a high correlation coefficient value, i.e. a reasonably good match - though not the correct one. This is caused by a "low frequency component" in the ring width curve that creates a special shape or pattern. When the two curves lay almost at the right overlapping position then these patterns seem to go into each other and the correlation coefficient then gives quite a high value. This phenomenon is sometimes described as "there is a lot of autocorrelation in the ring width curve" or that "the curve has autocorrelation". All of this is caused by the fact that a wide ring is often the neighbour of another wide ring which eventually makes up a certain overall shape or pattern for a specific ring width curve. A mathematician might say that a ring width is not independant of the adjacent ring width. |
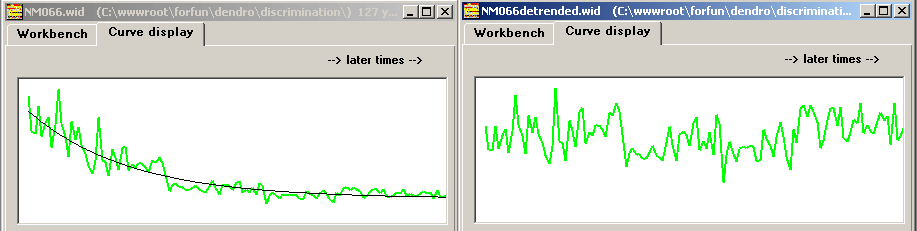

| After detrending the NM066 curve its autocorrelation function seems to indicate better chances for successful crossdating. I.e. the correct matching position (1995) stands out and there is no competing alternative. |
|
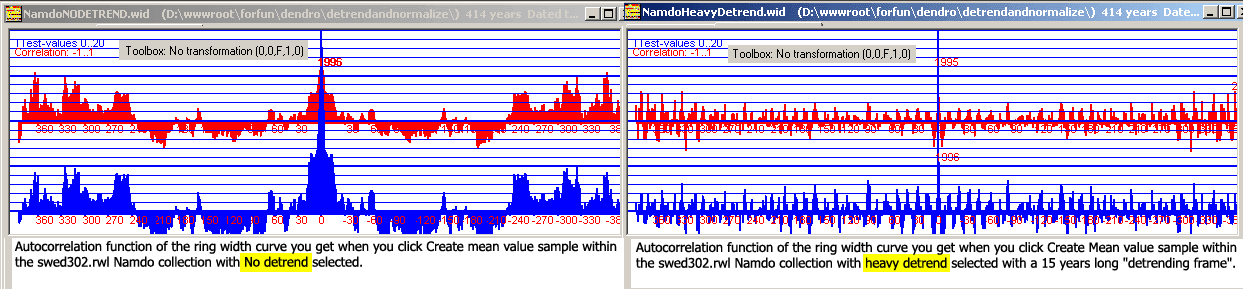
A CDendro collection has a button "Create Mean value sample". When you click this button a mean ring width value sample is created.
Its normalized values are then created as a mean of the normalized data within all the individual members of the collection.
A mean value ring width curve is also created after the individual members' ring width curves have been detrended. There is a small
frame under the button where you can specify which detrending method to use. After creating the mean value sample you may save it as a .wid file.
Depending on the detrending method used you get a little bit different looking mean value ring width curves also with a little bit different autocorrelation curves. |
|
With no detrending at all before calculating the ring width mean values, we then get quite an ugly autocorrelation curve from it.
The curve signals that using that mean value ring width curve for crossdating implies a high risk for incorrect results.
|
|
The "least autocorrelation" for a mean value ring width curve is achieved when the Heavy detrend method is applied to the collection members before the mean
value ring widths are calculated.
|
|
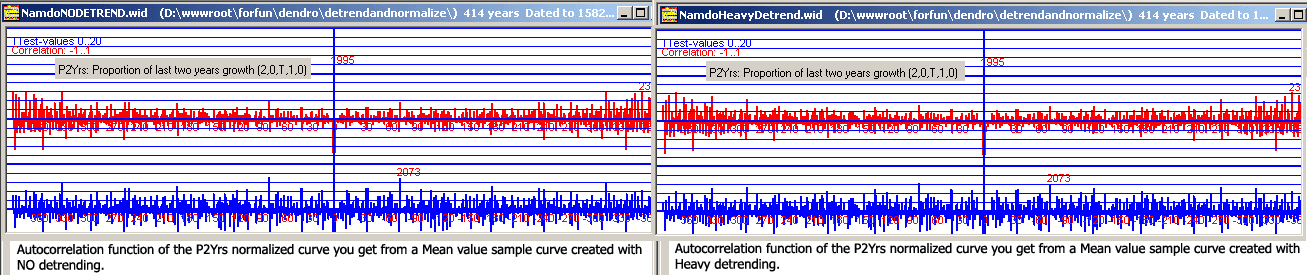
After normalization, we see little difference between autocorrelation curves created from normalized ring width curves based on mean values from detrended or from not detrended curves.
So we might question what the autocorrelation curves from ring width mean values actually tell us.
It is anyhow a bad idea to ignore the necessity of detrending when calculating ring width mean value curves! With high "sample depth" (i.e. many trees within the collection and many tree rings for each year - sometimes called high "replication") detrending has a limited influence on the mean value curve shape, but with lower sample depth (i.e. only a few tree rings for certain years) detrending will be more important for creating good mean value curves and for correct crossdating. |
|
To make good crossdating you need a good match (this usually means a T-value near to or above 6) which is also properly discriminated - i.e. there should be no competing match! To achieve good discrimination you have to "remove the autocorrelation" from your curves. This can be done by detrending your curves or by normalizing them or by doing it both ways. When crossdating, we recommend using several methods of normalization to achieve trustable crossdating results, see example below! |
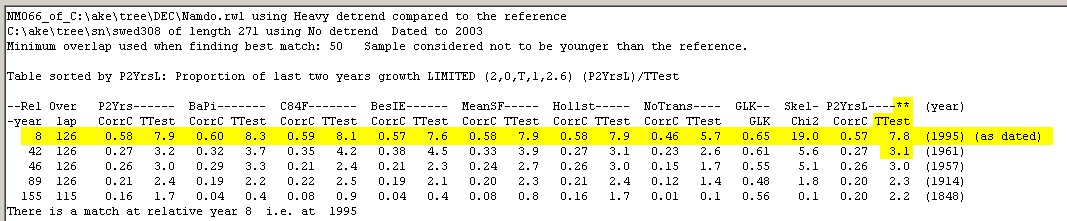
| The diagram above shows the result from crossdating a tree from the island of Nämdö towards the reference curve from Saltsjöbaden on the mainland. The best results are sorted according to the T-values from the P2YrsL method which here gives a relation of 7.8/3.1 between the best and the next best T-values - this means a good discrimination. |